A joint team led by Prof. Wei Xie from the School of Life Sciences at Tsinghua University, Dr. Xukun Lu from Institute of Women, Children and Reproductive Health at Shandong University, and Dr. Yu Zhang from Institute of Reproduction and Development at Fudan University unraveled how DNA methylation is re-established after implantation in mammals. Their study, titled “Reprogramming of H3K36me2 guides lineage-specific post-implantation de novo DNA methylation” was published in Nature Cell Biology on November 11, 2025. This study systematically mapped the dynamic reprogramming of H3K36me2 during early mouse embryogenesis and revealed how H3K36me2 reprogramming coordinates with DNMT3A/3B to regulate lineage-specific post-implantation de novo DNA methylation.
DNA methylation plays critical roles in gene expression, genomic imprinting, and X chromosome inactivation, the defect of which causes developmental defects, infertility, and diverse human diseases. As a key carrier of epigenetic memory, DNA methylation is faithfully inherited during cell division in somatic cells. However, during mammalian gametogenesis and early embryogenesis, DNA methylation is dynamically reprogrammed to facilitate the parental-to-zygotic transition and reset the developmental clock. In the mammalian life cycle, three major waves of de novo DNA methylation establishment occur during spermatogenesis, oogenesis, and post-implantation embryonic development. While DNA methylation establishment in spermatogenesis and oogenesis are strictly regulated by histone methylation H3K36me2 and H3K36me3, respectively [1,2], the molecular basis for post-implantation DNA methylation re-establishment remains poorly understood.
In this study, the researchers first systematically profiled H3K36me2 in mouse oocytes, pre- and post-implantation embryos. They found that the transcriptional state of oocytes profoundly affected the interconversion between H3K36me2 and H3K36me3. In transcriptionally inactive oocytes, H3K36me2 was more enriched in gene bodies. After fertilization, paternal H3K36me2 was lost in 1-cell embryos while maternal H3K36me2 is gradually diminished by the 8-cell stage. After zygotic genome activation (ZGA), H3K36me2 is de novo established in a “two-step” manner. It first “seeds” at enhancers during pre-implantation development, then “spreads” across the genome (except on the inactive X chromosome) in post-implantation embryos.
The researchers then asked whether H3K36me2 is required for DNA methylation re-establishment after implantation. They established a mouse model in which NSD1, the major H3K36me2 methyltransferase, was catalytically mutated. Loss of H3K36me2 severely impaired DNA methylation re-establishment in extra-embryonic tissues, but only mildly affected embryonic tissues. Mechanistically, DNMT3A recognizes and depends strictly on H3K36me2/3 via its PWWP domain (accurate reader), whereas DNMT3B can methylate DNA in an H3K36me2/3-independent manner (leaky reader). Therefore, in embryonic tissues, the highly expressed DNMT3B compensated for the loss of H3K36me2, ensuring global methylation establishment. Nonetheless, H3K36me2 is required for proper methylation and silencing of methylation-prone promoters, including those of germline genes. The researchers further demonstrated that DNA methylation valleys (DMVs), where developmental genes are enriched, are protected from aberrant DNA methylation through PRC1/H2AK119ub1-mediated H3K36me2 exclusion.
Interestingly, DNMT3B is only lowly or not expressed in gametes. DNMT3A is the major de novo methyltransferase in sperm and oocyte. It is proposed that DNMT3A, a precise H3K36me2/3 reader, may be employed to accurately methylate genomic imprints, allele-specific DNA methylation that controls parental-specific gene expression, in gametes. Collectively, this study uncovers H3K36me2 reprogramming in mouse early development, including its two-step establishment process, and reveals its intriguing roles in guiding lineage- and locus-specific de novo DNA methylation and gene regulation after implantation. The work also highlights distinct dependencies of DNMT3A and DNMT3B on H3K36me2/3 in methylation establishment. This study deepens our understanding of mechanisms underlying de novo DNA methylation establishment in different biological processes (spermatogenesis, oogenesis, and post-implantation embryogenesis). These results will shed light on the fundamental mechanisms underlying infertility and development of better precise epigenome-editing tools.
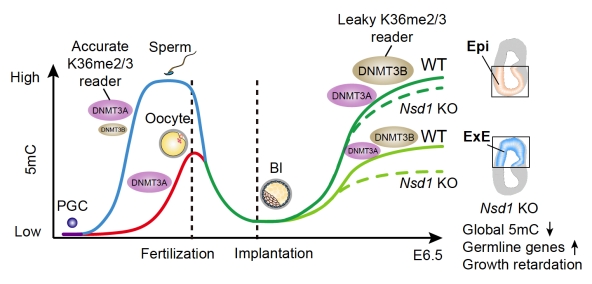
Figure 1. H3K36me2 coordinates with DNMT3A/3B to regulate post-implantation DNA methylation re-establishment in a lineage-specific manner
Prof. Wei Xie from the School of Life Science at Tsinghua University, Dr. Xukun Lu from Institute of Women, Children and Reproductive Health at Shandong University, and Dr. Yu Zhang from Institute of Reproduction and Development at Fudan University are the corresponding authors of this work. Xukun Lu and Lijuan Wang (Tsinghua University) are co-first authors. Bofeng Liu, Xiaoyu Hu, Zhengmao Wang, Ling Liu, Guang Yu, Lijun Dong, Feng Kong, and Qiang Fan made important contributions to this study. This work was supported by the National Key R&D Program of China, the National Natural Science Foundation of China, the Natural Science Foundation of Shanghai, the THU-PKU Center for Life Sciences. Prof. Wei Xie is a recipient of an HHMI International Research Scholar award and is a New Cornerstone Investigator. The authors also acknowledge the assistance of the Tsinghua University Laboratory Animal Center, the Biomedical Core Facility sequencing platform, and the computing platform.
Paper link: https://www.nature.com/articles/s41556-025-01805-8
References
1. Shirane, K., et al., NSD1-deposited H3K36me2 directs de novo methylation in the mouse male germline and counteracts Polycomb-associated silencing. Nat Genet, 2020. 52(10): p. 1088-1098.
2. Xu, Q., et al., SETD2 regulates the maternal epigenome, genomic imprinting and embryonic development. Nat Genet, 2019. 51(5): p. 844-856.
